Topic: Mitochondrial genome convergences
Most likely, mitochondria have a single evolutionary origin, but that doesn't mean they are immune to convergence...
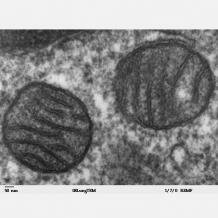 Current evidence points to mitochondria having a single origin as once free-living aerobic bacteria that became endosymbionts of the earliest eukaryotes. As part of the “surrender” to this symbiotic association, a substantial proportion of the bacterial genome was either lost or exported to the host genome, which is of course located in the nucleus. The net result was that the mitochondrion became wholly dependent on the host, while mitochondrial processes also affect a wide range of developmental and physiological events in the host. For example, mitochondrial genes have a direct effect on pollen development in plants and also seem to be important for animal reproduction and spermatogenesis. This leads to very interesting, and perhaps unexpected, convergences. Did you know…?
Current evidence points to mitochondria having a single origin as once free-living aerobic bacteria that became endosymbionts of the earliest eukaryotes. As part of the “surrender” to this symbiotic association, a substantial proportion of the bacterial genome was either lost or exported to the host genome, which is of course located in the nucleus. The net result was that the mitochondrion became wholly dependent on the host, while mitochondrial processes also affect a wide range of developmental and physiological events in the host. For example, mitochondrial genes have a direct effect on pollen development in plants and also seem to be important for animal reproduction and spermatogenesis. This leads to very interesting, and perhaps unexpected, convergences. Did you know…?
MutS protein homologues
During DNA replication and recombination, errors may arise that need to be fixed if DNA function is to be maintained. Not surprisingly, the processes involved in such DNA mismatch repair are highly conserved. So it is that Mut proteins constitute the major component of the prokaryotic repair system. In the bacterium Escherichia coli, the MutS protein recognises mismatched bases and binds the mutated DNA. This is then nicked by the endonuclease MutH after activation by MutL. Within the eukaryotic nucleus, MutS homologues (the MSH proteins) are found and they also serve to repair DNA mismatches and direct recombination functions. However, less is known about their role in mitochondria.
 A mitochondrial MutS homologue termed MSH1 has been reported in a cnidarian (the soft coral Sarcophyton glaucum). This protein is special in that it possesses a homing endonuclease domain at its C-terminus, a feature lacking from other MutS homologues. It has been suggested that this is the result of an evolutionary fusion of MutS with MutH. Recently, a similar mitochondrial gene fusion has been discovered in higher plants such as soybean, tomato and maize. Intriguingly, the MutS domain of the plant protein is homologous with that of the cnidarian protein, but the endonuclease domains fused to them are so different that convergent evolution in response to similar selective pressures has evidently occurred. This convergence could be related to mitochondrial genome size. In higher plants, the mitochondrial genome varies dramatically in size and structure. The mitochondrial genomes of Cnidaria are equally divergent. Such changes in genome configuration might have required different sets of proteins for DNA maintenance. There is, however, another similarity between plants and corals, and this is their sessile lifestyle. Sessility imposes reproductive constraints on an organism and should in principle affect its mating system. Many plants exhibit gynodioecy, where hermaphroditic and female (male-sterile) individuals occur in a population. Mating systems in corals are strikingly similar. It has been suggested that mitochondrial MSH1 activity might affect sex determination. It will be intriguing to see whether the novel Msh1 is absent from the mitochondrial genomes of motile cnidarians.
A mitochondrial MutS homologue termed MSH1 has been reported in a cnidarian (the soft coral Sarcophyton glaucum). This protein is special in that it possesses a homing endonuclease domain at its C-terminus, a feature lacking from other MutS homologues. It has been suggested that this is the result of an evolutionary fusion of MutS with MutH. Recently, a similar mitochondrial gene fusion has been discovered in higher plants such as soybean, tomato and maize. Intriguingly, the MutS domain of the plant protein is homologous with that of the cnidarian protein, but the endonuclease domains fused to them are so different that convergent evolution in response to similar selective pressures has evidently occurred. This convergence could be related to mitochondrial genome size. In higher plants, the mitochondrial genome varies dramatically in size and structure. The mitochondrial genomes of Cnidaria are equally divergent. Such changes in genome configuration might have required different sets of proteins for DNA maintenance. There is, however, another similarity between plants and corals, and this is their sessile lifestyle. Sessility imposes reproductive constraints on an organism and should in principle affect its mating system. Many plants exhibit gynodioecy, where hermaphroditic and female (male-sterile) individuals occur in a population. Mating systems in corals are strikingly similar. It has been suggested that mitochondrial MSH1 activity might affect sex determination. It will be intriguing to see whether the novel Msh1 is absent from the mitochondrial genomes of motile cnidarians.
Mitochondrial gene loss
In four major eukaryotic lineages (animals, most fungi, chlamydomonad green algae and Apicomplexa, a large group of alveolate protists), all ribosomal protein genes have independently been lost from the mitochondrial genome. In contrast, flowering plants exhibit high variability in mitochondrial genome content. The common ancestor of angiosperms possessed 40 known mitochondrial protein genes. While the most basal angiosperms have retained all of them, others have significantly reduced their mitochondrial genome content. This reduction is convergent, having occurred, for example, in the orders Asparagales (in the monocot genus Allium, the onions), Poales (in the bogbutton Lachnocaulon) and Geraniales (genus Erodium), where almost complete or complete loss of ribosomal protein and succinate dehydrogenase (sdh) genes has occurred.
 Other losses are even more dramatic. Independently, several eukaryotes have lost their entire mitochondrial genome. Given the main metabolic role of mitochondria is in respiration, this loss is linked with the adoption of an anaerobic or parasitic lifestyle. Examples include the Microsporidia, obligate intracellular parasitic fungi, and a wide range of protists, such as the anaerobic trichomonads and rumen-dwelling ciliates (where the non-respiring, agenomic mitochondrion is referred to as a hydrogenosome).
Other losses are even more dramatic. Independently, several eukaryotes have lost their entire mitochondrial genome. Given the main metabolic role of mitochondria is in respiration, this loss is linked with the adoption of an anaerobic or parasitic lifestyle. Examples include the Microsporidia, obligate intracellular parasitic fungi, and a wide range of protists, such as the anaerobic trichomonads and rumen-dwelling ciliates (where the non-respiring, agenomic mitochondrion is referred to as a hydrogenosome).
Interestingly, an analysis of mitochondrial gene loss has also led to the suggestion that the genome similarities observed between all major lineages of plastids (the cell organelles of plants and algae involved in photosynthesis and compound storage) are the result of convergence. If plastids had a single evolutionary origin, surely they should be more similar to each other than to a mitochondrion, but this does not seem to be the case. The mitochondrial genome of the heterotrophic flagellate protist Reclinomonas americana, which contains 97 genes, shows remarkable similarities with the genomes of primary plastids. Fifty-two of the 111 genes encoded in the plastid of the liverwort Marchantia are also found in Reclinomonas mitochondria, considerably more than what would be expected if gene loss was a random process. Hence, a constraint on gene loss might explain these genome similarities.
The cox2 gene
In other cases of mitochondrial gene loss, the function of the gene is still needed, so it has migrated (that is translocated) to the nuclear genome. One example is the mitochondrial respiratory gene cox2 (cytochrome c oxidase subunit II). This and its function was transferred to the nucleus during the diversification of legumes, a group of plants famous for their symbiotic association with nitrogen-fixing bacteria. Some legume species still contain intact mitochondrial and nuclear cox2 copies, which are transcribed at varying levels. It has been shown that inactivation of either one or the other copy has occurred multiple times in different ways (e.g. by complete gene loss or loss of transcript editing). As nuclear copies have been inactivated about as often as mitochondrial ones, no location seems to be selectively advantageous. Most likely, random mutations silenced either one or the other gene.
However, cox2 does provide an intriguing example of evolutionary convergence in another context. Functional gene transfer typically involves an entire gene being activated in the nucleus. Not so in the case of cox2 transfer in chlorophycean green algae. Here, cox2 was split into two genes, both of which then relocated to the nucleus.  In the chlorophycean Scenedesmus, the 3′ section was transferred, but the 5′ section remained in the mitochondrion, and vice versa in the lineage that contains Polytomella and Chlamydomonas. In apicomplexan protists, a strikingly similar cox2 split has occurred. Whilst this was initially interpreted as a case of lateral gene transfer, with apicomplexans taking up a green alga as an endosymbiont, a recent analysis of cox2 in dinoflagellates, the sister lineage to Apicomplexa, has provided evidence for an independent origin of the split cox2 gene in apicomplexans and green algae. This seemingly arcane example is important for two reasons: First, until recognised as convergent, it had been used as an argument to phylogenetically link the green algae to the alveolates. Second, just because an evolutionary event appears to be rare, one must not automatically assume it will only happen once.
In the chlorophycean Scenedesmus, the 3′ section was transferred, but the 5′ section remained in the mitochondrion, and vice versa in the lineage that contains Polytomella and Chlamydomonas. In apicomplexan protists, a strikingly similar cox2 split has occurred. Whilst this was initially interpreted as a case of lateral gene transfer, with apicomplexans taking up a green alga as an endosymbiont, a recent analysis of cox2 in dinoflagellates, the sister lineage to Apicomplexa, has provided evidence for an independent origin of the split cox2 gene in apicomplexans and green algae. This seemingly arcane example is important for two reasons: First, until recognised as convergent, it had been used as an argument to phylogenetically link the green algae to the alveolates. Second, just because an evolutionary event appears to be rare, one must not automatically assume it will only happen once.
TOM proteins
 As pointed out above, the mitochondrial genome is mostly concerned with respiration. A key aspect of its operation is the reception and transport of proteins across the outer and inner membranes, which is achieved by two protein complexes, TOM (translocase of the outer membrane) and TIM (translocase of the inner membrane). Some of the TOM proteins, such as TOM7, TOM22 and TOM40, are evidently very ancient – but not all of them. In animals and fungi, we find TOM20, which is absent in plants. What is found here, however, is a direct analogue, representing a convergent solution. Interestingly, this protein has a reversed configuration, with the C-terminus anchored to the outer membrane, whereas in animals and fungi it is the N-terminus. This recalls the remarkable convergence in insect olfactory and gustatory transmembrane protein receptors, with a convergent “opsin” that has the reversed configuration of true opsin. It remains to be established whether protists possess TOM20, have lost it or again have a convergent equivalent.
As pointed out above, the mitochondrial genome is mostly concerned with respiration. A key aspect of its operation is the reception and transport of proteins across the outer and inner membranes, which is achieved by two protein complexes, TOM (translocase of the outer membrane) and TIM (translocase of the inner membrane). Some of the TOM proteins, such as TOM7, TOM22 and TOM40, are evidently very ancient – but not all of them. In animals and fungi, we find TOM20, which is absent in plants. What is found here, however, is a direct analogue, representing a convergent solution. Interestingly, this protein has a reversed configuration, with the C-terminus anchored to the outer membrane, whereas in animals and fungi it is the N-terminus. This recalls the remarkable convergence in insect olfactory and gustatory transmembrane protein receptors, with a convergent “opsin” that has the reversed configuration of true opsin. It remains to be established whether protists possess TOM20, have lost it or again have a convergent equivalent.
Cite this web page
Map of Life - "Mitochondrial genome convergences"
https://mapoflife.org/topics/topic_440_mitochondrial-genome-convergences/
April 22, 2021

